|
New Features in 2020
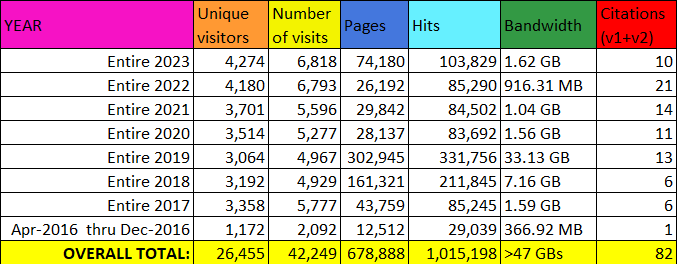
In 2016, we published the first version of our database in NAR; and since then, the database has shown value to the research community, generating over 1 million hits from 26,455 unique visitors. Our database has been increasingly referenced, further demonstrating its enduring impact on the field. This growing body of users stands to benefit from the latest developments on our website, which include an expanded library of data as well as improved functionality. Over the past three years, we have more than doubled the size of our database while adding several new features in ADPriboDB v2.0. For example, we now include the sites of ADP-ribosylation for nearly 48,000 database entries. This inclusion will facilitate new analyses, particularly those that seek to correlate sites of ADP-ribosylation with other protein modifications, such as phosphorylation. To facilitate this type of correlative analyses, we have developed a new interface integrating various post-translational modification data.
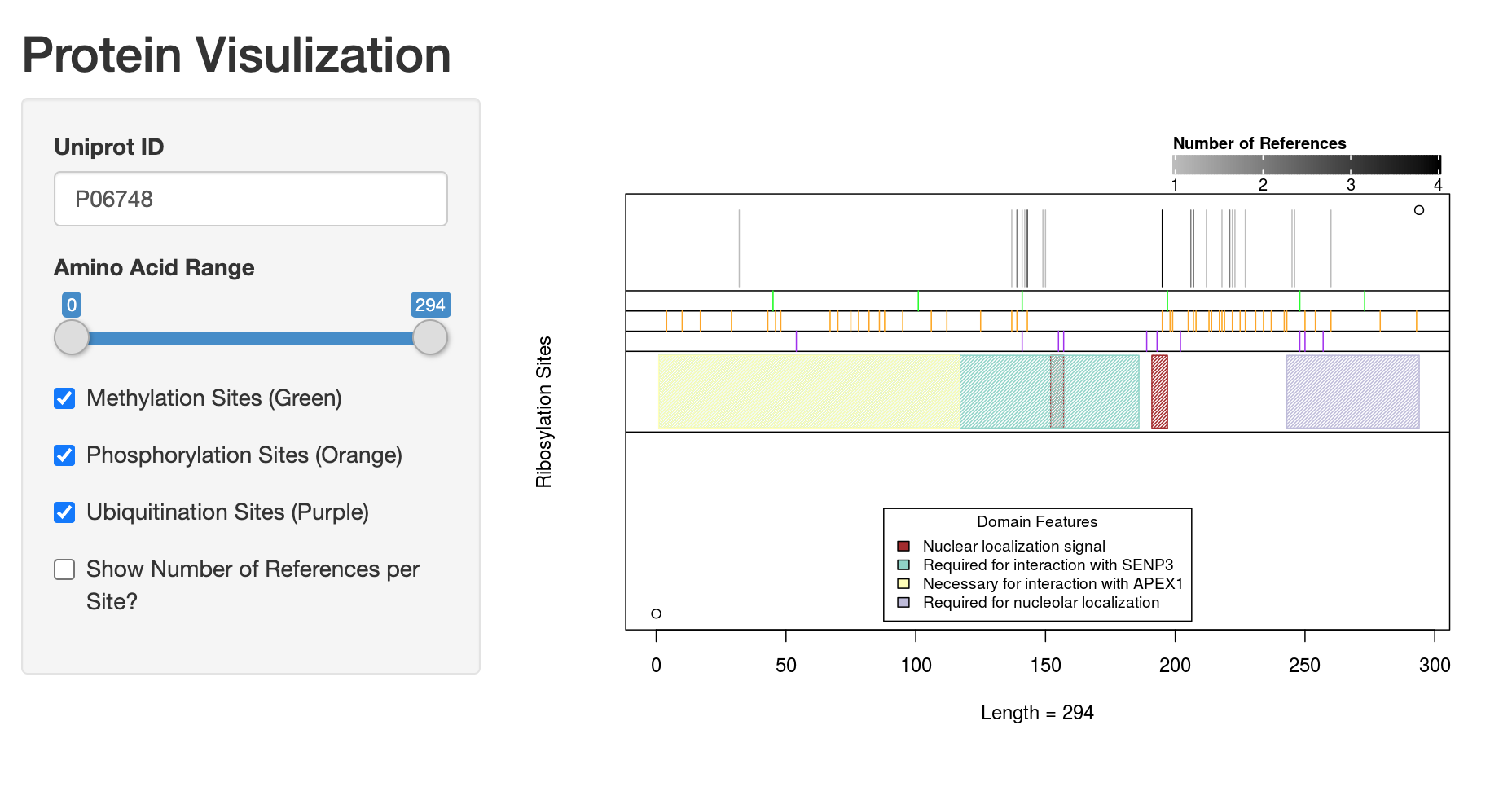
An Expanded Library of Data Increased database size The latest iteration of our database comprises 48,346 entries, including 9,097 unique proteins, 31,926 sequences, and 14,839 unique sites across 610 papers. These entries include data derived from 41 species, compared with 28 species in the previous database. ADPriboDB v2.0 has more than doubled the proportion of its entries bearing information regarding the enzyme responsible for the ADP-ribosylation. Reflecting a greater availability of technology (e.g. proteome arrays), 65% of database entries include the information of enzyme-substrate specificity. Increased database speed To enhance the accessibility of this larger database size, we have also used the open-source PHP framework Xataface to enhance the speed of database loading. By saving requested pages into a cache, and subsequently displaying cached version of each page, this approach allows vast improvements in the speed of information loading and display. Availability of ADP-ribosylation Site Information Information regarding the location of ADP-ribosylation enables users to perform mechanistic analyses through mutagenesis. We have now provided site information for ~25% of database entries. Each entry in our database for which site information exists comes with additional data presented in the "Other Sites" entry, revealing other sites of ADP-ribosylation identified in that protein. Moreover, we now add an additional entry, "Other Publications," that provides the PMID's of other papers that have also identified that particular site as ADP-ribosylated. As a result, users can prioritize to pick these sites of high confidence for experimental analyses. New Graphical Interface Visualizing ADP-ribosylation sites To facilitate user-specific analyses of sites, we have integrated our database with a Shiny-based web application that allows users to interactively visualize proteins with site information available. A graphical representation is shown with vertical lines corresponding to locations of modified sites. To help identify ADP-ribosylation sites of high confidence for further experimental analyses, the number of publications that identified a given modified site is now provided. Users are free to toggle zoom settings to visualize crowded regions at a single amino acid level. Below is an example of such a visualization for the NPM1 protein. Correlative analyses with other post-translational modifications Given that different protein modifications interact with one another, ADPriboDB v2.0 allows users to compare ADP ribosylation sites listed for a given protein to sites of phosphorylation, ubiquitination, or methylation from dbPTM (Huang et al., Nucleic Acids Research 2016; PMID: 26578568). We have now provided an easy visual correlation of ADP-ribosylation sites with phosphorylation, ubiquitination, and methylation. This feature pairs nicely with external links provided on our database to other databases, such as Phosphosite, that can direct users to more detailed information regarding these other modifications. |